We are studying Bioinformatics and Computational genomics
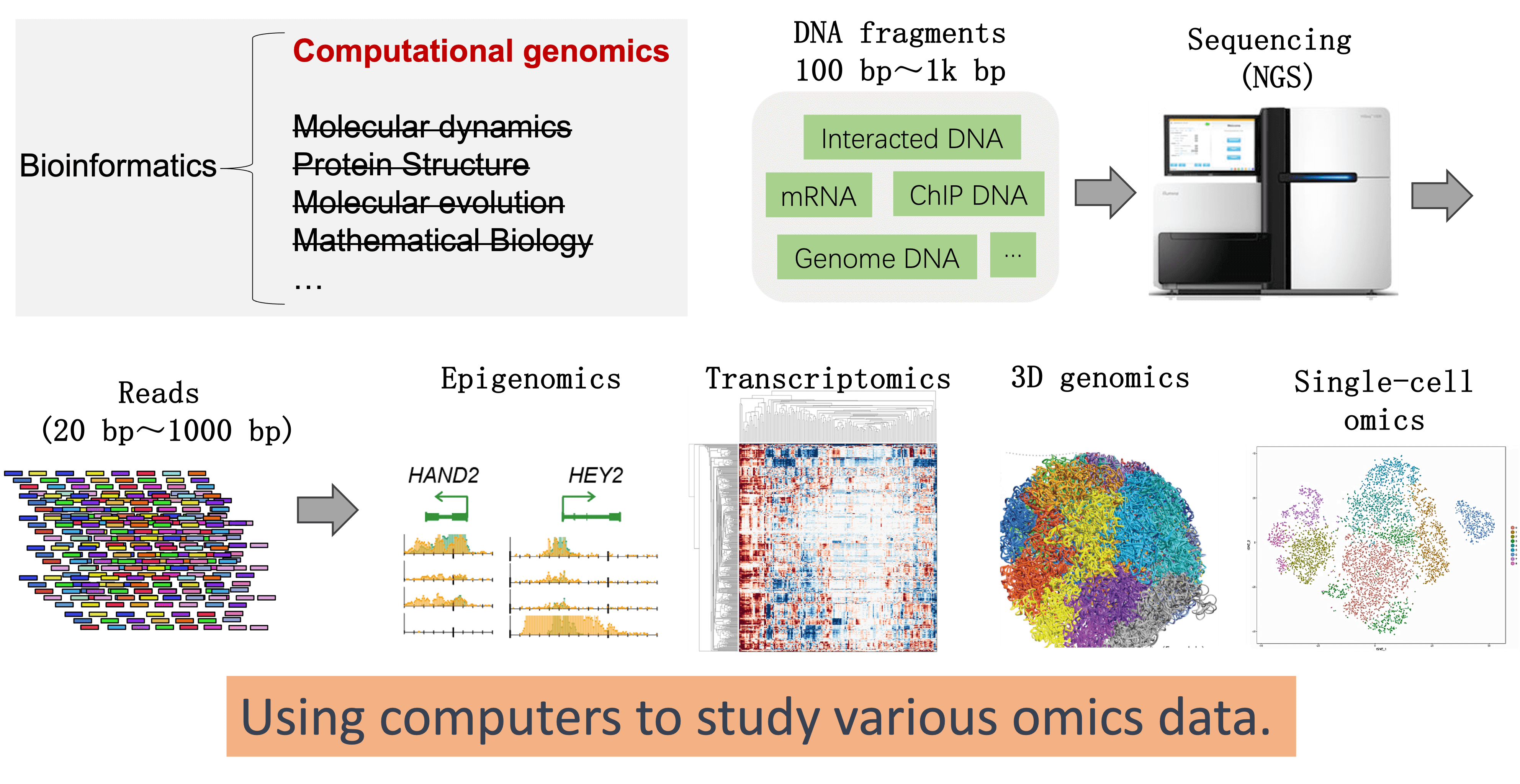
Bioinformatics is an interdisciplinary field that involves the application of computational techniques to store, manage, analyze, and interpret biological data. Computational genomics is a subset of bioinformatics that specifically focuses on the analysis of genomic data.
The characteristics of this laboratory
- This lab is contructed with the idea of both wet-lab and dry-lab. A “wet lab” refers to a traditional laboratory setting where experiments are conducted using biological or chemical materials. A “dry lab” refers to a laboratory setting where computational and analytical work is performed using computers and software tools.

- The main differences from the 100% wet lab:
- Data-driven research, where the data obtained is authentic.
- Requires the development of new methods and tools, cannot solely rely on existing tools.
- The main differences from the 100% dry lab:
- Guided by life science questions (not computer problem-solving),
- Focusing on the latest sequencing technologies.
- Does not require highly specialized computer knowledge, with an emphasis on applications.
- Both programming and biomedical knowledge are needed.
- Emphasis on collaboration and interdisciplinary approaches.
- Available for wet lab facilities, allowing for wet-lab work as support.
Research Topics
This lab has a primary focus on g next-generation sequencing (NGS) technologies such as ChIP-seq/CUT&TAG, Hi-C/Hi-ChIP/ChIAPET, RNA-seq/PROseq/GROseq, ATAC-seq, Bisulfite-seq, and single-cell sequencing. We primarily developing software, algorithms, and databases related to these NGS technologies. We also leverages multi-omics big data mining to explore biomedical questions. The main research keywords (until now) are summaried as the following wordcloud picture:
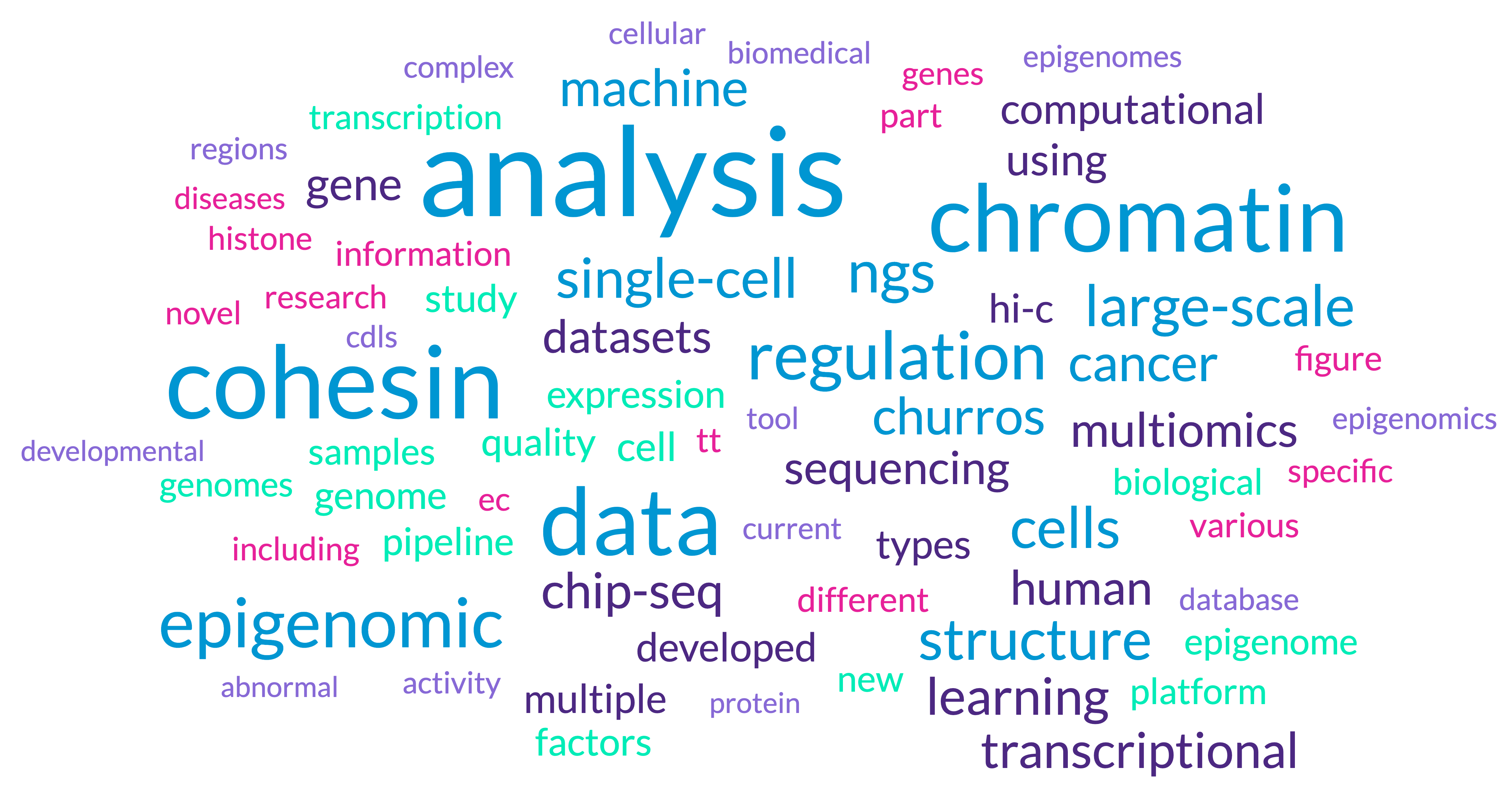
Our topics can also be summaried as:
- Epigenetic Genomics (e.g., ChIP-seq, ATAC-seq)
- 3D Genomics and Chromosome Architecture (e.g., Hi-C, ChIA-PET)
- Transcriptional Regulatory Mechanisms (Cohesin protein, cis-regulatory module)
- Multi-omics Integration (Large-scale multiomics analysis)
- Disease Genomics (e.g., CdLS genetic disorder, Cancer Genomics)
- Bioinformatics Software and Database Development
- Machine Learning/Deep Learning
- Single-cell Genomics
Two types of research projects
- Develop new methods/tools for NGS data:
- Software, Algorithms, or Databases.
- Addressing the Challenges in Genomic Data Analysis:
- Command-line (CLI), Web-based (Web), or Graphical User Interface (GUI).
- Require Programming and Interdisciplinary Skills.
- Do not rely on datasets.
- Purely dry experiments.
- Big Data Mining for Biomedical Research:
- Public Data, or Collaborative Research with Novel Data, or Integration of Wet and Dry Labs.
- Large-Scale Multi-omics Data in Human Diseases.
- Genetic Disorders, or Cancer genomics, or Other Diseases.
- Data-Driven Research.
- Purely dry experiments, or Predominantly dry Work with Wet-experiments Support
Current research projects
1. Develop a computational system for large-scale multiomics analysis.
2. Cohesin’s functions and related diseases.
3. Cancer research based on multiomics.
4. Re-analysis of high-quality public datasets.
5. Investigate histone modifications and transcriptional factors in single cell level
6. Investigate gene regulatory network by scCRISPR.
Programing Language
Linux/Shell, Python, R, JavaScript, Docker, HTML/CSS, Django, MySQL, C++, Java.
Facilities
- CPU server 256线程处理器,2048GB运行内存,200TB raid6硬盘
- GPU server 128线程处理器,512 GB 运行内存,两块RTX4090显卡,100TB Raid5硬盘
- HDD server 32线程处理器,128GB运行内存,600TB raid5 硬盘
- Workstation (Linux, MacOS, Windows)
- Benches, clean benches, cell incubators, and other basic wet lab equipment.
- (Shared within our department) Advanced equipment for all commonly used molecular biological experiments.