Research Introduction

Index
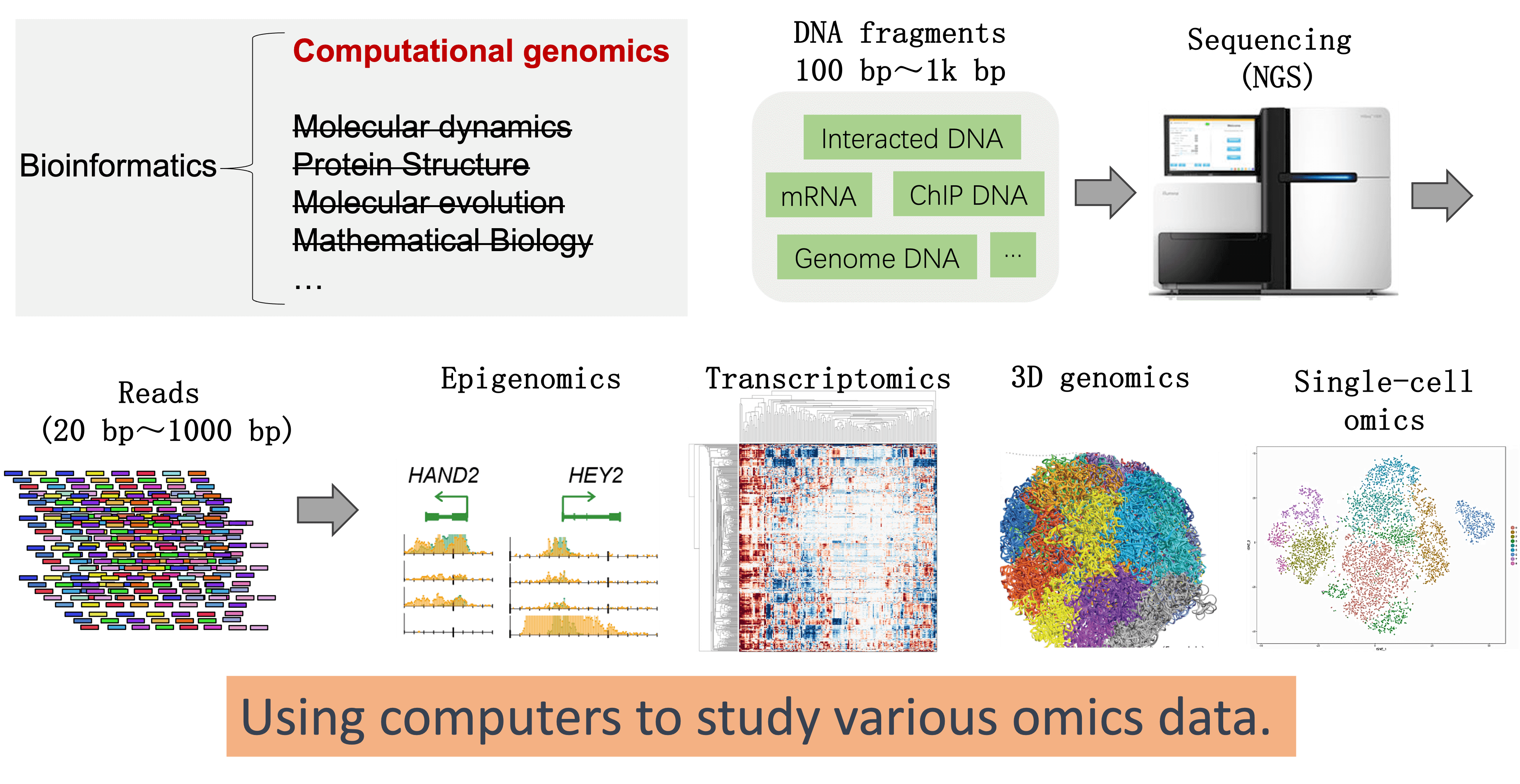
Bioinformatics and computational genomics
Transcriptional Regulation and Functional Genomics
The characteristics of this laboratory
Bioinformatics and Computational Genomics
Bioinformatics is an interdisciplinary field that involves the application of computational techniques to store, manage, analyze, and interpret biological data. Computational genomics is a subset of bioinformatics that specifically focuses on the analysis of genomic data.
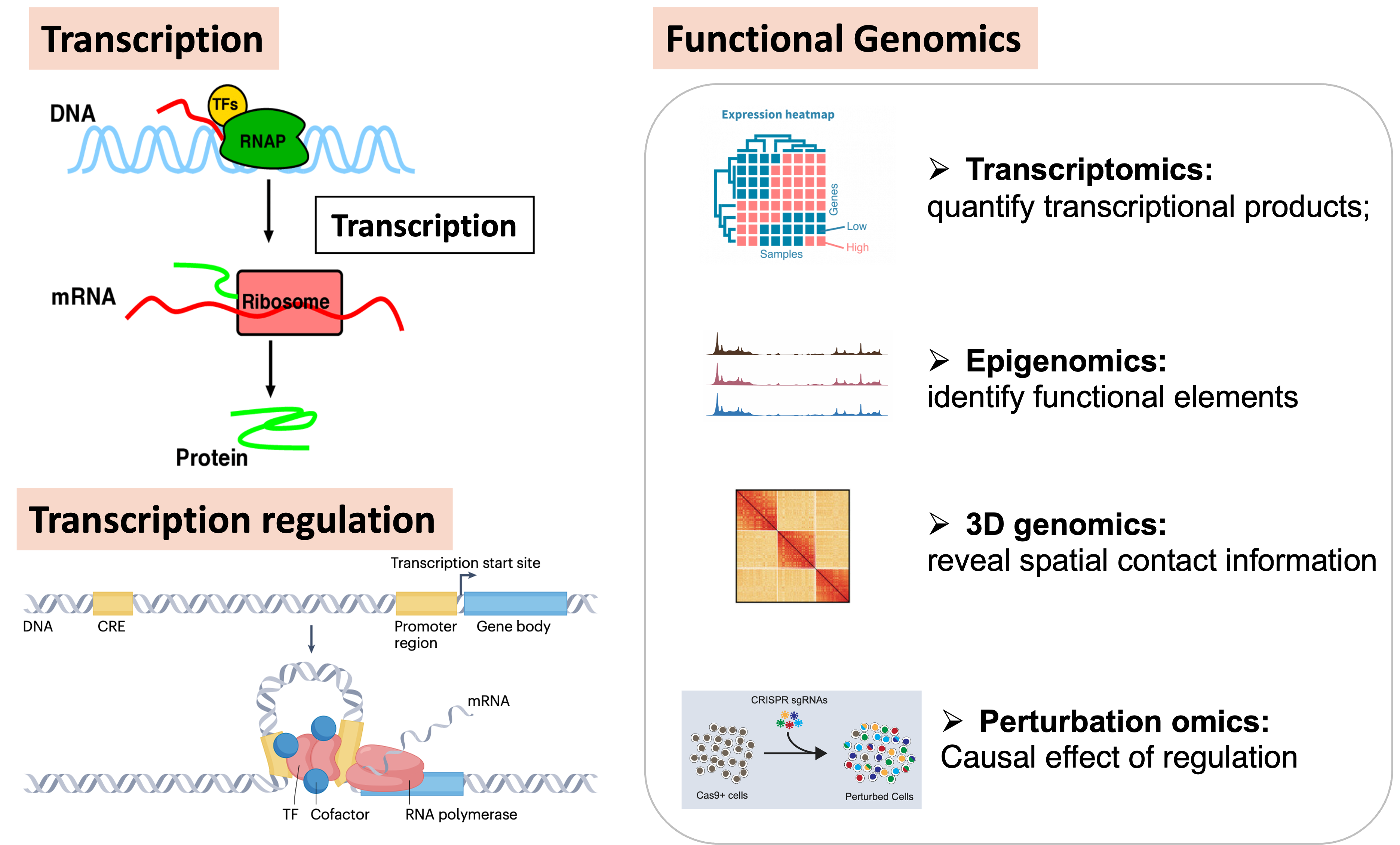
Transcriptional Regulation and Functional Genomics
Transcription is the most critical regulatory node in gene expression. Functional genomics research on transcriptional regulation plays a guiding role in understanding cellular activities and human diseases. In eukaryotes, transcriptional function depends on the intricate three-dimensional structure of chromosomes within the nucleus, forming multi-layered gene regulatory networks by enriching cis-regulatory elements for transcriptional regulators.
Currently, transcription can be described from multiple perspectives, including epigenomics, 3D genomics, transcriptomics, and perturbation omics. However, due to the complexity of integrating multidimensional and heterogeneous data, multi-omics joint analysis of transcriptional regulation faces numerous bioinformatics challenges.
Our research group engages in bioinformatics and functional genomics studies based on next-generation sequencing, aiming to:
- Develop robust and efficient bioinformatics methods for multi-omics joint analysis of transcriptional regulation.
- Integrate multi-omics data-driven research to uncover unexplored regulatory mechanisms in transcription.
This will significantly enhance the efficiency of transcriptional regulation research and accelerate scientific understanding of transcriptional activities and human diseases.
Research Topics
This lab has a primary focus on g next-generation sequencing (NGS) technologies such as Epigenomics (ChIP-seq/CUT&TAG,ATAC-seq, Bisulfite-seq), 3D genomics (Hi-C/Hi-ChIP/ChIAPET), Transcriptomics (RNA-seq/PROseq/GROseq), Perturbation omics (CRISPR screening, Perturb-seq), at the bulk and single-cell level.
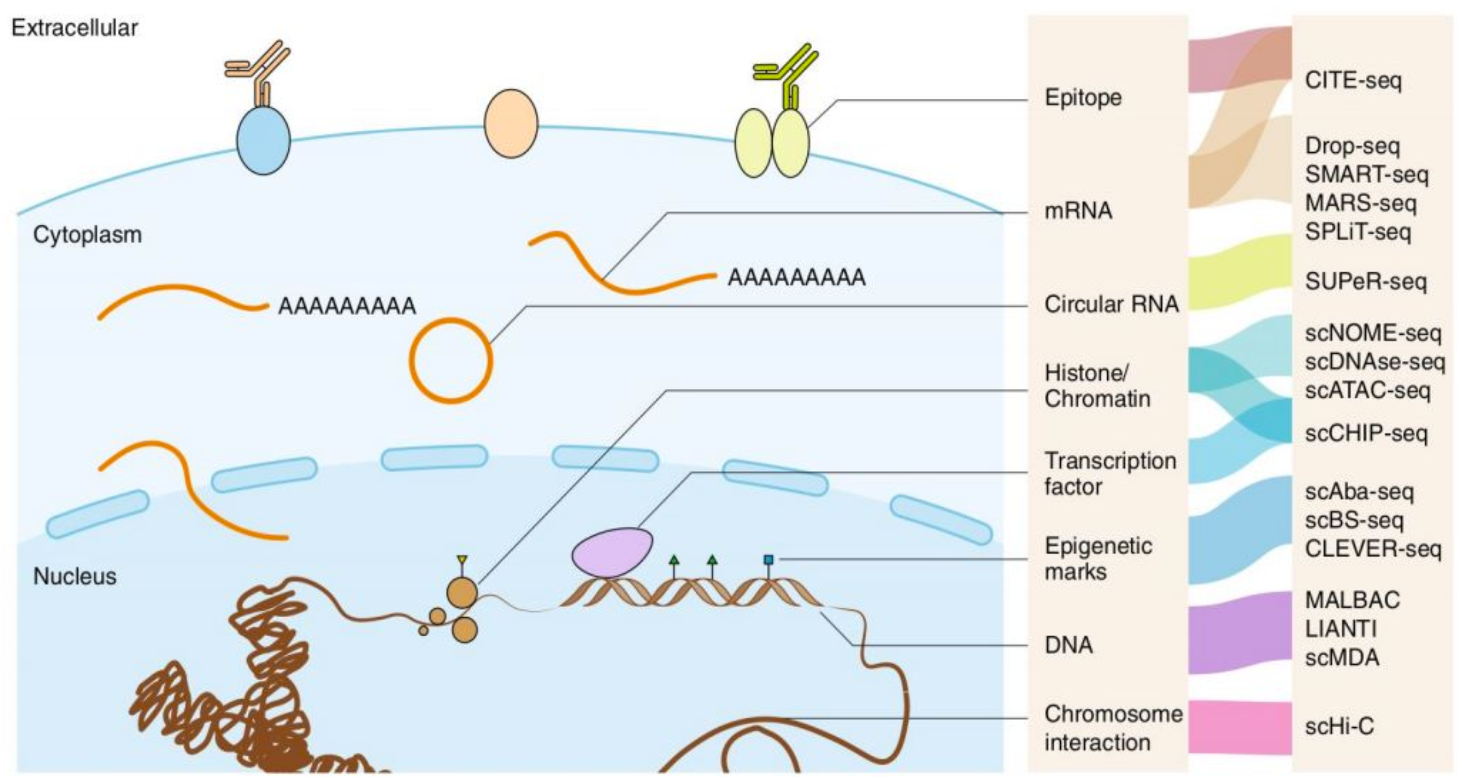
We primarily developing software, algorithms, and databases related to these NGS technologies. We also leverages multi-omics big data mining to explore biomedical questions. The main research keywords (until now) are summaried as:
- Epigenetic Genomics (e.g., ChIP-seq, ATAC-seq)
- 3D Genomics and Chromosome Architecture (e.g., Hi-C, ChIA-PET)
- Transcriptional Regulatory Mechanisms (Cohesin protein, cis-regulatory module)
- Multi-omics Integration (Large-scale multiomics analysis)
- Disease Genomics (e.g., CdLS genetic disorder, Cancer Genomics)
- Bioinformatics Software and Database Development
- Machine Learning/Deep Learning
- Bulk and Single-cell Multiomics
Some of the research projects are list as following:
- Develop a computational integrating system for large-scale multiomics analysis.
- Local correlation between two epigenomic tracks
- Decipher gene regulatory machinery from the silencing of transcriptional factors.
- Investigate the temporal transcriptional regulation by time-series multi-omics data.
- Integrate single-cell and bulk sequencing data to analyze histone modification and transcriptional factors.
- Use multiomics and Transformer to impute ChIP-seq.
- The functional relationship between G4,cohesin and chromatin structure.
- Investigate histone modifications and transcriptional factors in single cell level
- Cohesin’s functions and related diseases.
- Cancer research based on multiomics.
Highlighting Aspects
We always focus on the following aspects in our lab

The “Two types of purpose”:
- Develop new methods/tools for NGS data:
- Software, Algorithms, or Databases.
- Addressing the Challenges in Genomic Data Analysis:
- Command-line (CLI), Web-based (Web), or Graphical User Interface (GUI).
- Require Programming and Interdisciplinary Skills.
- Do not rely on datasets.
- Purely dry experiments.
- Big Data Mining for Biomedical Research:
- Public Data, or Collaborative Research with Novel Data, or Integration of Wet and Dry Labs.
- Large-Scale Multi-omics Data in Human Diseases.
- Genetic Disorders, or Cancer genomics, or Other Diseases.
- Data-Driven Research.
- Purely dry experiments, or Predominantly dry Work with Wet-experiments Support
The characteristics of this laboratory
- This lab is contructed with the idea of both wet-lab and dry-lab. A “wet lab” refers to a traditional laboratory setting where experiments are conducted using biological or chemical materials. A “dry lab” refers to a laboratory setting where computational and analytical work is performed using computers and software tools.

- The main differences from the 100% wet lab:
- Data-driven research, where the data obtained is authentic.
- Requires the development of new methods and tools, cannot solely rely on existing tools.
- The main differences from the 100% dry lab:
- Guided by life science questions (not computer problem-solving),
- Focusing on the latest sequencing technologies.
- Does not require highly specialized computer knowledge, with an emphasis on applications.
- Both programming and biomedical knowledge are needed.
- Emphasis on collaboration and interdisciplinary approaches.
- Available for wet lab facilities, allowing for wet-lab work as support.
Lab Facilities
- CPU server 256 threads CPU,2048GB memory,200TB raid6 harddisk
- GPU server 128 threads CPU,512 GB memory,two RTX4090 GPU,100TB Raid5 harddisk
- HDD server 32 threads CPU,128GB memory,600TB raid5 harddisk
- Workstation (Linux, MacOS, Windows)
- Benches, clean benches, cell incubators, and other basic wet lab equipment.
- (Shared within our department) Advanced equipment for all commonly used molecular biological experiments.
Programing Language
Linux/Shell, Python, R, JavaScript, Docker, HTML/CSS, Django, MySQL, C++, Java.